💡 This study employs a two-way, two-sample Mendelian randomization (MR) approach to investigate the potential causal relationship between gut microbiota and two outcomes of influenza: pneumonia without influenza and influenza pneumonia.
📌 Utilizing extensive meta-analysis data from the MiBioGen Alliance and GWAS data from the FinnGen consortium, the study identifies causal associations between specific gut microbiota and influenza infection, shedding light on potential preventive and therapeutic strategies for influenza.
📌 The findings highlight the gut-lung axis as a crucial factor in the interaction between gut microbiota and influenza, paving the way for further exploration of this intricate relationship.
📍 Methods:
Mendelian Randomization (MR):A two-way, two-sample MR approach was applied to assess the potential causal connection between gut microbiota and influenza.
Utilized GWAS data from the MiBioGen Alliance and FinnGen consortium to estimate single-nucleotide polymorphisms (SNPs) and conduct MR analyses.
Employed Inverse Variance Weighted (IVW), MR Egger, and Weighted Median (WM) analyses to estimate and summarize SNPs.
📍 Key Findings:
Associations with Influenza:
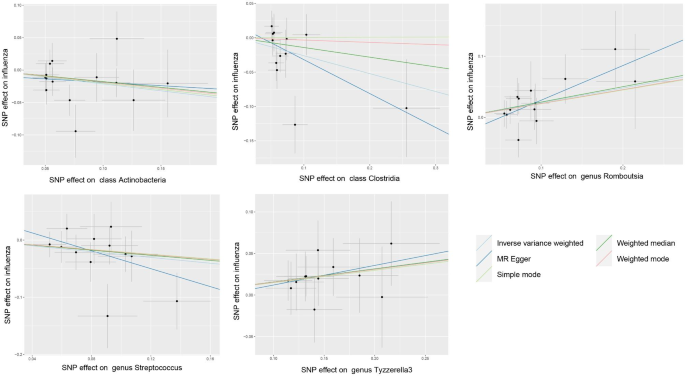
IVW analysis revealed significant associations between influenza infection and five bacterial taxa, with seven gut microbiota causally related to influenza infection. WM analysis largely supported IVW results, confirming the causal relationship between specific gut microbiota and influenza occurrence.
Associations with Influenza Pneumonia: Seven bacterial taxa exhibited a significant association with influenza pneumonia.
Gut-Lung Axis Interaction: The study suggests a causal relationship between certain gut microbiota and influenza, emphasizing the gut-lung axis. SCFA metabolism, particularly the influence of short-chain fatty acids (SCFAs), was identified as a potential mechanism in the gut-lung axis interaction.
Specific Bacterial Clusters: Identified potential protective effects against influenza infection by Actinobacteria and Clostridia, involved in SCFA metabolism. 𝘉𝘢𝘤𝘪𝘭𝘭𝘪 𝘢𝘯𝘥 𝘚𝘵𝘳𝘦𝘱𝘵𝘰𝘤𝘰𝘤𝘤𝘶𝘴 demonstrated bidirectional causal relationships with influenza.
New Bacterial Associations: Discovered potential links between influenza and specific gut bacterial groups such as 𝘙𝘰𝘮𝘣𝘰𝘶𝘵𝘴𝘪𝘢, 𝘛𝘺𝘻𝘻𝘦𝘳𝘦𝘭𝘭𝘢3, 𝘋𝘦𝘧𝘭𝘶𝘷𝘪𝘪𝘵𝘢𝘭𝘦𝘢𝘤𝘦𝘢𝘦, 𝘢𝘯𝘥 𝘉𝘢𝘳𝘯𝘦𝘴𝘪𝘦𝘭𝘭𝘢, warranting further investigation.
📌 SCFA metabolism emerges as a vital mechanism influencing the interaction between gut microbiota and influenza. Potential probiotic additions like 𝘊𝘭𝘰𝘴𝘵𝘳𝘪𝘥𝘪𝘶𝘮 , with protective effects against influenza and 𝘪𝘯𝘧𝘭𝘶𝘦𝘯𝘻𝘢 𝘱𝘯𝘦𝘶𝘮𝘰𝘯𝘪𝘢, are highlighted. Bidirectional causal relationships with specific bacterial clusters emphasize their potential as indicators of influenza and infection severity.
Link to the article : https://bmcinfectdis.biomedcentral.com/articles/10.1186/s12879-023-08706-x