The rapid rise of antibiotic resistance, coupled with the slow discovery of new antibiotics, poses a significant threat to global health. We truly require new antibacterial medicines since infections are become more and more resistant to drugs.
Phage lysins have grown into desirable alternatives because they break down the cell walls of bacteria and lead them to die quickly. However, the enormous amount of labor required for experimental screening stands in the way of the identification and validation of new lysins.
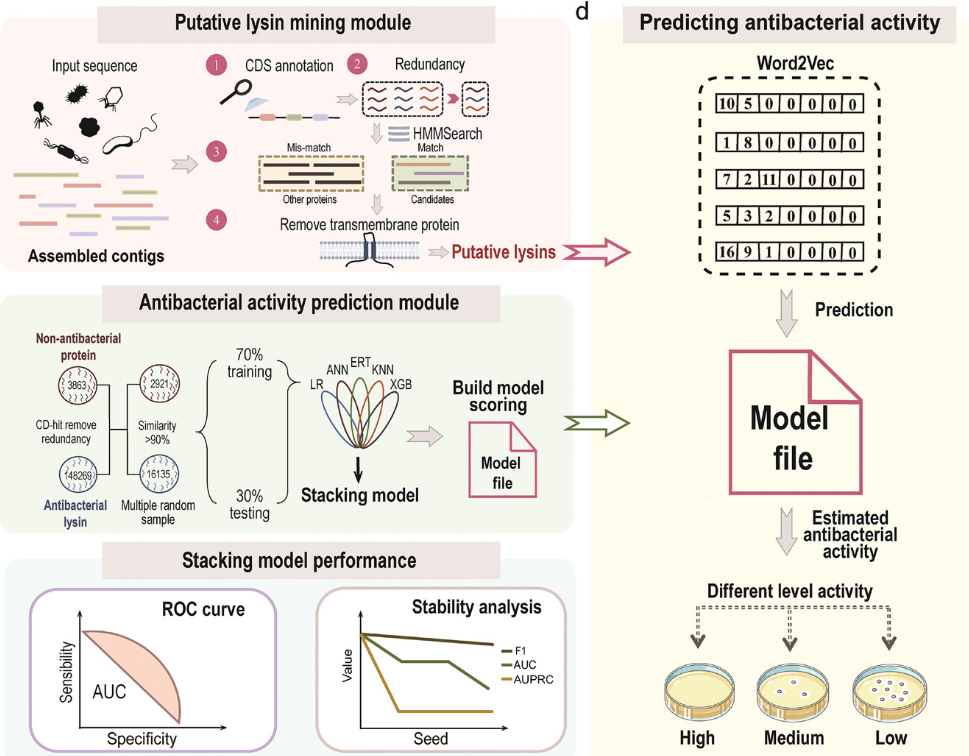
In this study, researchers provide DeepLysin, the first integrated software package that mines the massive genomic “dark matter” for new antimicrobial lysins by artificial intelligence. This novel strategy seeks to accelerate the identification of novel antibacterial proteins, offering a possible remedy for the growing problem of antibiotic resistance.
Methods:
- Software Engineering: The two main modules that make up DeepLysin are antibacterial activity prediction and lysin mining.
With the exception of transmembrane proteins, the lysin mining module annotates coding sequences, eliminates duplications, and uses HMMsearch to predict lysins.
To evaluate possible lysins, the antibacterial activity prediction module use a stacking model that combines a number of machine learning methods.
- Validation through experimentation: A panel of bacterial strains was used to experimentally validate seventeen lysins that were chosen at random. Mice models for bacteremia and wound infections were used in conjunction with in vitro tests to evaluate the effectiveness of lysins.
- Analyzing Data and Training Models: Protein sequences obtained from bacteria and phages were deduplicated and annotated. Using methods like Word2vec, features were retrieved, and a two-layer stacking model was trained to forecast antibacterial activity. The models’ performance was assessed through the use of metrics such as AUC, MCC, ACC, F1 score, and AUPRC.
- Testing both in vitro and in vivo: Time-kill tests, minimum inhibitory concentration (MIC) determination, and plaque formation assays were used to validate the antibacterial activity of lysins. In mouse models, in vivo efficacy was evaluated by measuring survival rates, bacterial load, and histological changes.
Key Results:
- DeepLysin Formation : A unified software package called DeepLysin was created to use AI to search large genome reservoirs for new antimicrobial phage lysins.
The program consists of two modules: one for predicting antibacterial activity and the other for mining lysin.
The software uses a stacking model that accurately predicts antibacterial activity and separates protein characteristics. - Validation through experimentation: After testing and synthesising seventeen new lysins, seven of them showed notable antibacterial activity. Among the lysins, LLysSA9 demonstrated remarkable efficacy, outperforming the best-in-class substitute lysin.
- Efficacy in Vivo and In Vitro: Excellent antibacterial activity was demonstrated by LLysSA9 against a variety of Staphylococcus aureus strains, including methicillin-resistant strains. LLysSA9 markedly decreased the bacterial load and increased survival rates in mouse models of bacteremia and skin wound infections.
- Artificial Intelligence in Drug Discovery: The study showed how artificial intelligence may be used to sift through vast amounts of genomic data, find, and assess novel antibacterial protein candidates. The combination of computational and experimental methods used by DeepLysin offers an innovative approach for the search for antibacterial drugs.
Link to the study : https://tinyurl.com/5n853c6h