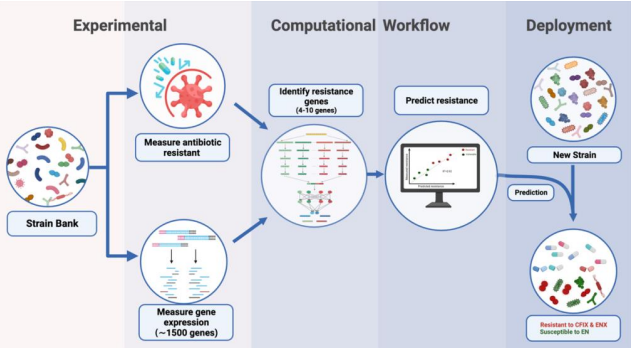
Antimicrobial resistance (AMR) poses a significant global health threat, necessitating precise identification and management strategies. This study explores a novel approach using transcriptomics and machine learning to predict antibiotic resistance profiles in pathogens, aiming to enhance treatment efficacy and combat AMR.
Methods:
The study employs a transcriptomics-based approach, focusing on gene expression patterns correlated with antibiotic resistance. Machine learning algorithms are utilized to analyze large datasets, identifying optimal models for predicting resistance. Feature reduction techniques specific to transcriptomic data are developed to enhance model performance and interpretability.
Key Findings:
- Transcriptomics in AMR Prediction: Gene expression serves as a robust indicator of resistance, enabling accurate prediction across various antibiotics.
- Machine Learning Integration: ML algorithms, including Random Forest and SVM, significantly improve predictive accuracy by learning complex resistance patterns from transcriptomic data.
- Importance of Dataset Quality: High genetic diversity and a sufficient number of resistant samples are crucial for training effective models, highlighting the need for comprehensive strain banks.
- Feature Reduction Challenges: Novel methods for feature reduction in transcriptomic data are essential to mitigate overfitting and enhance model generalizability.
- Clinical Implications: Automated ML frameworks offer scalable solutions for real-time resistance profiling, potentially revolutionizing AMR surveillance and personalized treatment strategies.
This study underscores the potential of integrating transcriptomics and machine learning to address AMR challenges effectively. By advancing predictive accuracy and scalability in resistance profiling, these methods pave the way for tailored antibiotic therapies and improved patient outcomes in the fight against AMR.
Link to the article : https://tinyurl.com/6fy27mtt