The gut virome, comprising primarily bacteriophages with some eukaryotic and archaeal viruses, plays a crucial role in maintaining gut homeostasis and influencing the immune system. Despite its importance, a comprehensive understanding of the gut virome in IBD, particularly across diverse populations, is limited.
This study aimed to analyze the gut virome in a Chinese cohort of IBD patients and healthy controls, identifying viral signatures associated with IBD and exploring their relevance across different populations.
Methods
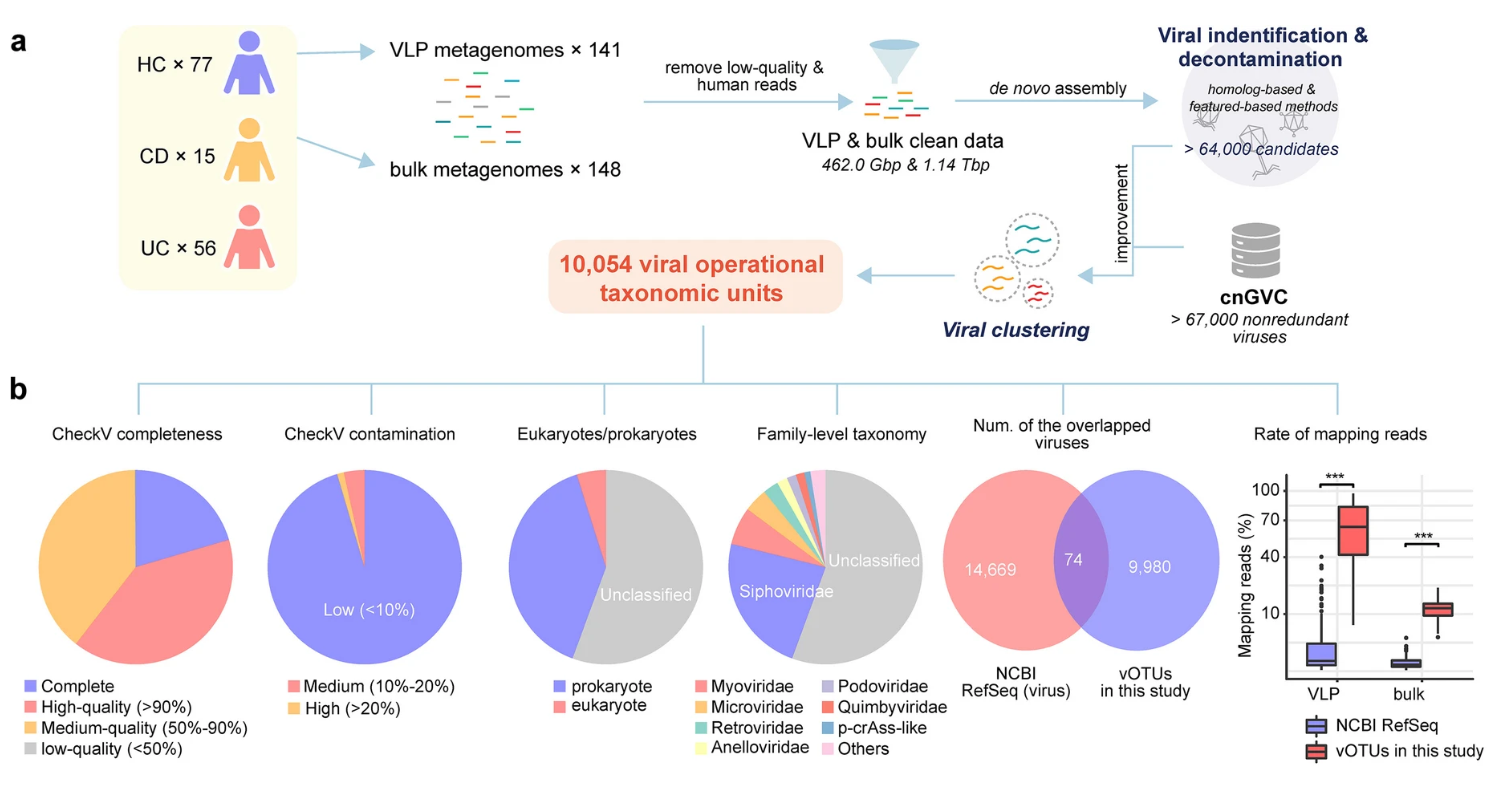
A total of 148 participants were included in the study, comprising 71 IBD patients (15 with Crohn’s disease and 56 with ulcerative colitis) and 77 healthy controls. The gut virome was analyzed using two approaches: viral-like particle (VLP) enrichment and bulk virome sequencing of fecal samples. An integrated gut virus catalog tailored to the IBD virome was established to identify viral signatures. Alpha- and beta-diversity analyses, as well as differential abundance analyses, were conducted to characterize the gut virome. Fecal virome transplantation (FVT) experiments were performed to verify the role of these viral signatures in modulating experimental colitis in mouse models.
Key Findings
Alterations in the Gut Virome: Identified 139 differentially abundant viral signatures in IBD patients, including elevated phages targeting Escherichia, Klebsiella, Enterococcus_B, Streptococcus, and Veillonella species. IBD-depleted phages targeting Prevotella, Ruminococcus_E, Bifidobacterium, and Blautia species were noted.
Consistency Across Populations: The identified viral signatures showed high consistency across diverse populations in Europe and the USA, indicating their broad relevance in the disease context.
Experimental Validation: Fecal virome transplantation experiments demonstrated that IBD-characterized viruses could modulate experimental colitis in mouse models.
Potential Biomarkers: Identified potential viral biomarkers for prognosis and therapy in IBD patients, providing a foundation for further exploration of viromes in related conditions.
Eukaryotic and Prokaryotic Virome Diversity: Increased eukaryotic virome richness and evenness in IBD patients, particularly in the Retroviridae and Genomoviridae families. Contrasting results in prokaryotic virome diversity between VLP and bulk datasets, with VLP showing increased diversity and bulk datasets showing decreased diversity in IBD patients.
Functional Insights: Enrichment of temperate phages in IBD patients, associated with opportunistic pathogens like Escherichia and Klebsiella, which are known to induce inflammation. Depletion of crAss-like and Quimbyviridae viruses, potentially implicating a loss of viral functions related to polysaccharide metabolism in the gut virome of IBD patients.
This study provides a comprehensive analysis of the gut virome in IBD patients, revealing significant alterations and identifying viral signatures with potential diagnostic and therapeutic implications. The findings underscore the importance of utilizing complementary sequencing techniques and viral reference databases to fully explore the diversity of viruses within the human gut.
Link to the study : https://tinyurl.com/4ncs7fch