The human microbiome, the community of microorganisms living in our bodies, plays a crucial role in our overall health. Researchers have often identified correlations between certain bacterial groups and various health conditions, but understanding the precise genetic and ecological factors driving these associations remains challenging.
A novel approach called reverse ecology aims to identify genetically and ecologically distinct bacterial populations that have adapted to specific niches within the human body. This study investigates whether genome-wide selective sweeps (GWSS) are a common adaptive mechanism within the human microbiome and how these sweeps influence bacterial population structures and associations with human health.
Methods
To test the hypothesis that GWSS create ecologically differentiated populations, the researchers employed several steps:
- Identification of Vertical Descent: They differentiated clonally inherited genome portions from recombined ones to identify vertical descent among closely related genomes.
- Theoretical Modeling: A model was used to estimate the likelihood of genome clusters on long phylogenetic branches arising from GWSS.
- Mapping Metagenomes: The researchers mapped metagenomic data onto these clusters to check for missing intermediate genotypes and to confirm whether GWSS clusters occupy distinct ecological niches.
- Testing for Selection and Associations: They looked for signatures of selection and differential associations with human phenotypes as proxies for different ecological conditions in the gut.
Key Findings
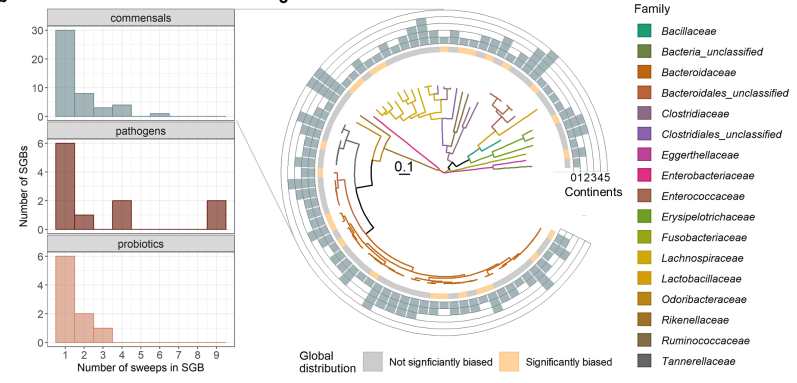
- Evidence of GWSS in Human Microbiome: Phylogenetic analysis of human microbiome isolates often showed a “broom” structure, indicating the presence of GWSS. This structure consists of a cluster of closely related strains (the broom head) on long phylogenetic branches (the broom handle).
- Ecological Differentiation: GWSS clusters were found to occupy different niche spaces within the human gut, indicating that these populations are ecologically differentiated.
- Global Spread of GWSS Clusters: GWSS clusters of gut commensals appear to spread globally in a manner similar to epidemics, although at a slower pace compared to pathogens. This slower spread allows for greater diversity within commensal GWSS clusters as they adapt to different environments.
- Association with Human Phenotypes: The study found that GWSS clusters are differentially associated with human phenotypes, suggesting that they can serve as precise markers for various health conditions. The mapping of GWSS clusters to metagenomes could be useful for identifying ecological differentiation and constructing bacterial co-occurrence networks.
- Potential for Experimental Models: Identifying GWSS clusters can help select appropriate bacterial isolates for experimental models or synthetic gut communities, potentially improving research on microbiome-related diseases and therapies.
This study highlights the significance of GWSS in shaping bacterial populations within the human microbiome. These sweeps create ecologically differentiated populations that can be traced and studied to better understand their roles in health and disease. By identifying these adaptive populations, researchers can develop more targeted approaches for studying microbiome functions and developing probiotic treatments. The findings suggest that GWSS are a fundamental force in microbiome evolution, with implications for understanding microbial adaptation and its impact on human health.
Link to the article : https://tinyurl.com/58uudfp5