𝘗𝘴𝘦𝘶𝘥𝘰𝘮𝘰𝘯𝘢𝘴 𝘢𝘦𝘳𝘶𝘨𝘪𝘯𝘰𝘴𝘢 is a notorious pathogen due to its high resistance to multiple antibiotics, complicating the treatment of infections. This study focuses on predicting the multi-drug resistance (MDR) phenotypes of 𝘗𝘴𝘦𝘶𝘥𝘰𝘮𝘰𝘯𝘢𝘴 𝘢𝘦𝘳𝘶𝘨𝘪𝘯𝘰𝘴𝘢 isolates through genotypic analysis, aiding in timely and effective antibiotic treatment.
Methods
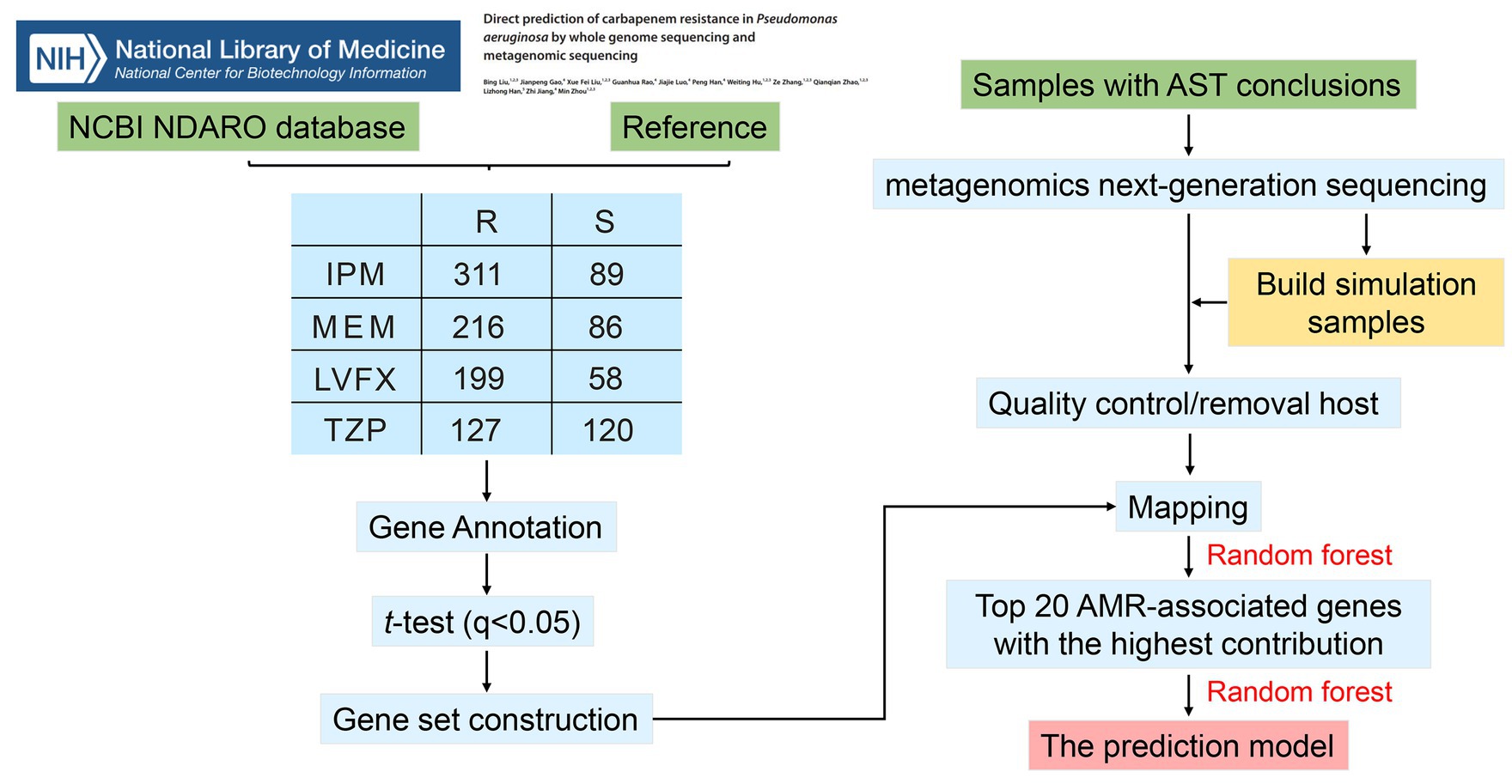
The study utilized whole genome sequencing (WGS) data from 494 𝘗𝘴𝘦𝘶𝘥𝘰𝘮𝘰𝘯𝘢𝘴 𝘢𝘦𝘳𝘶𝘨𝘪𝘯𝘰𝘴𝘢 isolates to identify antimicrobial resistance (AMR)-associated genes related to resistance against imipenem (IPM), meropenem (MEM), piperacillin/tazobactam (TZP), and levofloxacin (LVFX). By comparing gene copy number differences between resistant and sensitive strains, key AMR genes were identified. Metagenomics next-generation sequencing (mNGS) was then performed on 74 𝘗𝘴𝘦𝘶𝘥𝘰𝘮𝘰𝘯𝘢𝘴 𝘢𝘦𝘳𝘶𝘨𝘪𝘯𝘰𝘴𝘢 positive sputum samples, excluding one low-quality sample, to construct resistance prediction models.
Key Findings
1. AMR-associated Genes Identification:
– The study identified 93, 88, 80, and 140 AMR-associated features for IPM, MEM, TZP, and LVFX resistance, respectively.
2. Prediction Model Development:
– The top 20 AMR-associated features for each antibiotic were used to develop resistance prediction models using the random forest algorithm.
– These models demonstrated strong predictive performance, with areas under the curve (AUC) greater than 0.8 for all antibiotics.
3. Model Performance:
– The high AUC values indicate that the models reliably predict the resistance of 𝘗𝘴𝘦𝘶𝘥𝘰𝘮𝘰𝘯𝘢𝘴 𝘢𝘦𝘳𝘶𝘨𝘪𝘯𝘰𝘴𝘢 to IPM, MEM, TZP, and LVFX.
4. Significance of AMR Genes:
– Key genes such as ymdF, stbD, hcnA, and uspA play significant roles in resistance mechanisms, contributing to factors like biofilm formation, toxin-antitoxin systems, and stress response.
5. mNGS Utility:
– mNGS proved effective in rapidly predicting AMR by directly detecting resistance-associated genes, offering a quicker alternative to traditional drug susceptibility testing.
6. Challenges and Implications:
– The study highlighted challenges like high costs, host DNA contamination, and complex data interpretation associated with mNGS.
– Despite these challenges, the findings underscore mNGS’s potential in clinical settings for rapid and accurate prediction of antibiotic resistance in 𝘗𝘴𝘦𝘶𝘥𝘰𝘮𝘰𝘯𝘢𝘴 𝘢𝘦𝘳𝘶𝘨𝘪𝘯𝘰𝘴𝘢, guiding appropriate antibiotic therapy.
Conclusion
The study successfully demonstrated that mNGS could be used to predict the resistance of 𝘗𝘴𝘦𝘶𝘥𝘰𝘮𝘰𝘯𝘢𝘴 𝘢𝘦𝘳𝘶𝘨𝘪𝘯𝘰𝘴𝘢 to multiple antibiotics by identifying and utilizing AMR-associated genes. This approach provides a promising tool for the rapid detection of drug resistance in clinical settings, potentially improving the management of MDR infections.
Link to the study : https://tinyurl.com/ymuhvrxe