This study investigates the evolution of the gut microbiome across hominids, including diverse human populations and wild-living chimpanzees, bonobos, and gorillas. Through high-resolution metagenomic analyses, we identify taxonomic and functional changes, providing insights into the diverging trajectories of host-associated microbial communities.
Key Scientific Findings:
- Host-Specific Gut Microbiota:
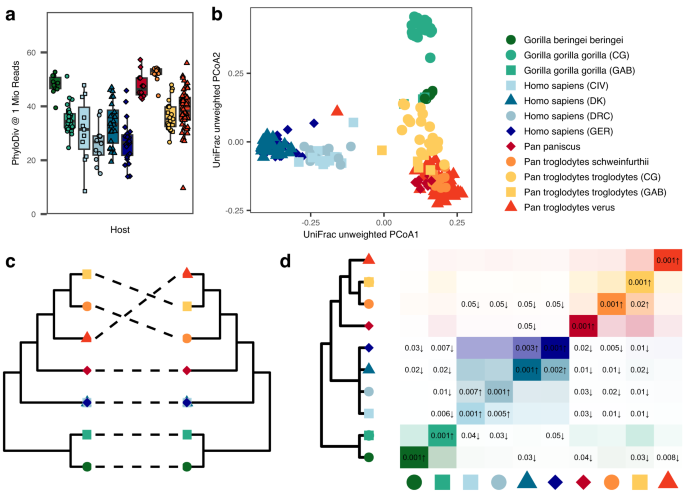
- Human-associated taxa are depleted in non-human apes, indicating the acquisition of novel microbial clades during host evolutionary divergence.
- Patterns of co-phylogeny between microbes and hosts are disrupted in humans, suggesting unique evolutionary dynamics.
- Diversity Loss in Human Populations:
- Pervasive loss of microbial diversity in human populations correlates with a high Human Development Index.
- Evolutionarily conserved clades show decreased abundance, potentially linked to industrialization and Western lifestyles.
- Functional Adaptations in European Populations:
- Functional traits related to aerobic respiration are enriched in European human populations, indicating microbial adaptation to high oxygen conditions.
- Introduction of novel microbes associated with industrialization leads to substantial community composition differences in European populations.
- Prevotella Depletion and Enterotype Dynamics:
- Prevotella, a conserved taxon across hominids, is depleted in European populations, suggesting departure from an evolutionary ancestral community state.
- Reduced abundance and diversity of Prevotella may serve as a biomarker for disease risk associated with Western lifestyles.
- Co-Phylogeny Patterns:
- Co-phylogeny analysis reveals clade-specific enrichments and depletions, with human-derived genomes less prevalent among co-phylogenetic groups.
- Traits related to reduced genome sizes, gene counts, and susceptibility to bile indicate microbial specialization in colonizing the hominid gut.
- Co-phylogeny analysis relies on metagenome-assembled genomes, potentially introducing biases, emphasizing the need for more robust genomic data.
- The study focuses on humans and African great apes, offering insights into recent adaptations but potentially neglecting broader-scale dynamics.
- Future research should explore horizontal gene transfer events and consider time series data for a comprehensive understanding of host-microbiome interactions.
Conclusion: This comprehensive analysis of the hominid gut microbiome provides valuable insights into the impact of evolutionary forces on microbial communities. Understanding the interplay between host evolution, environmental changes, and microbial adaptations enhances our knowledge of human-associated microbiota, offering potential avenues for personalized interventions in chronic inflammatory disorders.
Link to the study : http://tinyurl.com/4zj7az6s