💡 This study aimed to investigate the synergistic effects of host genes and gut microbes associated with CAD through integrative genomic analyses. This study sheds light on the role of gut microbes in CAD and provides potential biomarkers for CAD. The interplay between host genes and gut microbes associated with CAD through integrative genomic analyses is explored.
📍 Methods:
Fecal and blood samples were collected from 52 CAD patients and 50 healthy controls.
Amplicon sequencing of the 16S rRNA gene and transcriptomic sequencing were performed.
Differential analysis of the gut microbiota in CAD patients and controls was carried out.
Machine learning, specifically the Random Forest method, was employed to create a model for CAD patient classification using gut bacteria and host genes as biomarkers.
📍 Key Findings:
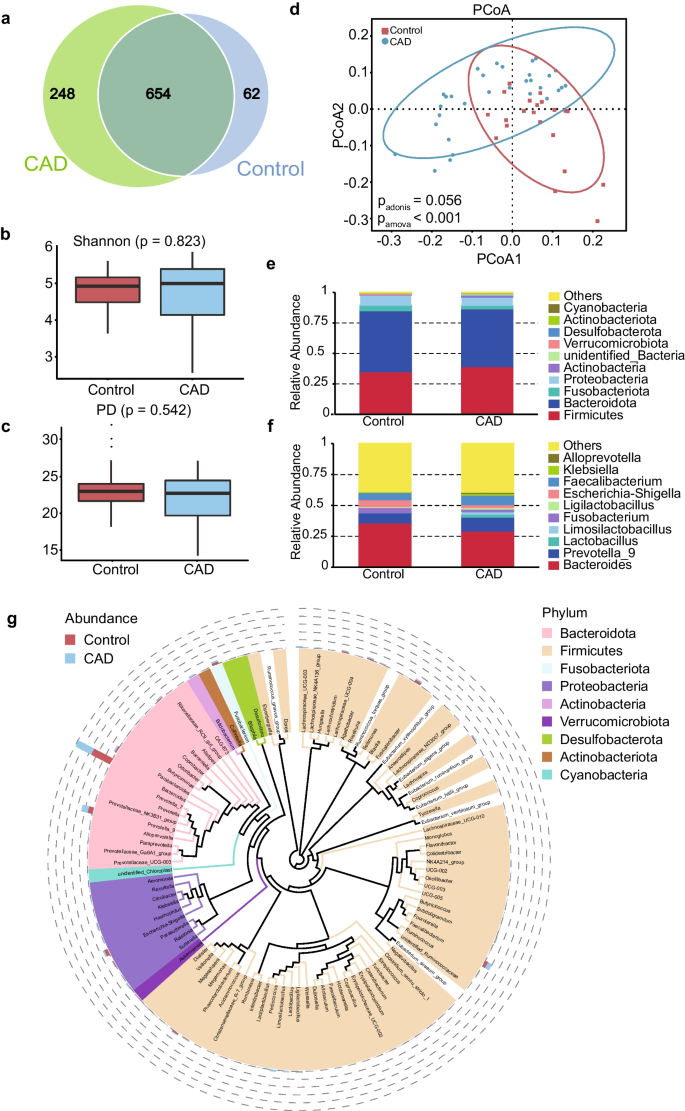
📌 Gut Microbial Dysregulation: By comparing the gut microbiota of CAD patients with healthy controls, they identified significant dysregulation in the gut microbial composition of CAD patients. Specifically, the depletion of certain genera, such as 𝘉𝘭𝘢𝘶𝘵𝘪𝘢, 𝘌𝘶𝘣𝘢𝘤𝘵𝘦𝘳𝘪𝘶𝘮, 𝘍𝘶𝘴𝘪𝘤𝘢𝘵𝘦𝘯𝘪𝘣𝘢𝘤𝘵𝘦𝘳 was observed, while other genera, including 𝘚𝘶𝘵𝘵𝘦𝘳𝘦𝘭𝘭𝘢 𝘢𝘯𝘥 𝘊𝘰𝘭𝘭𝘪𝘯𝘴𝘦𝘭𝘭𝘢, were enriched in CAD patients.
📌 Biomarker Discovery: Utilizing a Random Forest method, they developed a model that combined 20 bacterial and 30 gene biomarkers, achieving a high performance in distinguishing CAD patients from healthy controls with an area under the curve (AUC) of 0.92. This suggests the potential of these biomarkers for CAD diagnosis.
📌 Gut Microbe-Host Gene Interactions: Integration analysis revealed that the genus Fusicatenibacter, a gut microbe associated with CAD, was linked to the expression of CAD-risk genes, such as GBP2, MLKL, and CPR65. This interaction aligns with previous findings and provides insights into the underlying mechanisms of CAD.
📌 Immune-Related Pathways: CAD patients exhibited an upregulation of immune-related pathways, primarily associated with higher abundances of genera 𝘉𝘭𝘢𝘶𝘵𝘪𝘢, 𝘌𝘶𝘣𝘢𝘤𝘵𝘦𝘳𝘪𝘶𝘮, 𝘍𝘶𝘴𝘪𝘤𝘢𝘵𝘦𝘯𝘪𝘣𝘢𝘤𝘵𝘦𝘳, 𝘢𝘯𝘥 𝘔𝘰𝘯𝘰𝘨𝘭𝘰𝘣𝘶𝘴 . This suggests a potential link between the gut microbiota and immune responses in CAD.
📌 Diagnostic Potential: Gut microbiome-based diagnostic models demonstrated high accuracy, with an AUC of 93.9%. Combining gut microbiome data with clinical variables further improved the predictive power, yielding an AUC of 90.4%. These results emphasize the diagnostic potential of gut microbiota in CAD.
📌 Interferon Signaling Pathways: The study revealed a significant reduction in interferon signaling pathways in CAD patients, with a strong association between the depleted bacteria (𝘉𝘭𝘢𝘶𝘵𝘪𝘢, 𝘌𝘶𝘣𝘢𝘤𝘵𝘦𝘳𝘪𝘶𝘮, 𝘢𝘯𝘥 𝘍𝘶𝘴𝘪𝘤𝘢𝘵𝘦𝘯𝘪𝘣𝘢𝘤𝘵𝘦𝘳) and genes involved in interferon signaling. This suggests a potential mechanism by which gut microbes influence CAD.
📍 The study underscores the contribution of dysregulated gut microbes to CAD risk and their interaction with host genes. The identified gut microbes and interacting host genes hold promise as potential biomarkers for CAD. The findings emphasize the importance of understanding the role of the gut microbiota in CAD pathophysiology.
Link to the Article: https://tinyurl.com/3zekjj4w