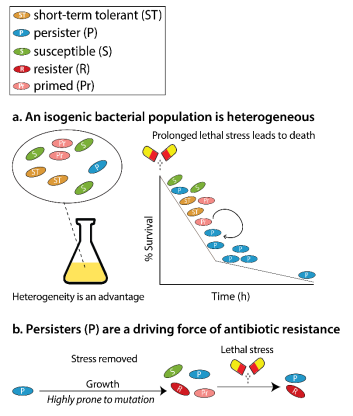
Antibiotic resistance poses a significant threat to global health, contributing to millions of deaths each year. A crucial factor in the recurrence of infections and the increased usage of antibiotics is the presence of a small subset of bacterial cells known as persisters.
Unlike antibiotic-resistant cells, persisters survive antibiotic treatment by entering a non-dividing state, thereby avoiding the lethal effects of antibiotics. This study explores the role of phenotypic diversity and epigenetic memory in the formation of persister cells and introduces the concept of “primed cells,” which are cells that prepare for antibiotic stress before encountering it. Understanding these mechanisms is vital for developing strategies to combat persistent infections and curb antibiotic resistance.
Key Findings
Phenotypic Diversity and Persister Formation:
- Bacterial populations exhibit significant fluctuations in gene expression, even under stable conditions. This phenotypic diversity allows some cells to enter a non-dividing state, becoming persisters that survive antibiotic exposure.
- Persister cells are not genetically distinct from the rest of the population but are characterized by their ability to withstand antibiotics due to their dormant state.
Primed Cells and Epigenetic Memory:
- The study identifies “primed cells” in E. coli, which are dividing cells prepared for antibiotic stress. These cells are more likely to become persisters and can pass this prepared state down through several generations via epigenetic memory.
- Primed cells are common across distant bacterial lineages, including Gram-positive bacterium Bacillus megaterium, indicating that this phenomenon is widespread in bacteria.
- Epigenetic memory in primed cells is transient, lasting for about seven generations.
Growth Phase and Persister Levels:
- The levels of persisters are higher in the transition and stationary phases compared to the log phase. In B. megaterium, persister levels increase by approximately two-fold in the transition phase and four-fold in the stationary phase compared to the log phase.
- The study challenges the previously proposed classification of Type I and Type II persisters, showing that primed cells are responsible for increased persister levels without distinct phenotypic differences.
Noise in Gene Expression and Bet-Hedging:
- Gene expression noise leads to phenotypic plasticity, enabling some cells to survive environmental stress through bet-hedging. This variability is critical for the survival of a subset of cells under lethal conditions.
- Primed cells and persisters are examples of how bacterial populations utilize phenotypic heterogeneity to withstand antibiotic stress.
Implications for Antibiotic Treatment:
- Understanding the formation of primed cells and their role in persister cell development can help in designing more effective antibiotic treatments.
- The findings suggest that targeting the mechanisms behind phenotypic diversity and epigenetic memory could reduce the prevalence of persisters and improve the efficacy of antibiotics.
Relevance and Importance
This study sheds light on the intricate mechanisms behind bacterial persistence, emphasizing the roles of phenotypic diversity and epigenetic memory in the survival of bacterial cells under antibiotic stress. By identifying primed cells and their contribution to persister formation, this research provides a new perspective on bacterial survival strategies. These insights are crucial for developing novel approaches to combat antibiotic resistance, improving treatment outcomes, and reducing the global burden of persistent bacterial infections. Understanding these mechanisms not only enhances our knowledge of bacterial physiology but also informs the development of therapies that can effectively target and eradicate persisters, ultimately helping to mitigate the rise of antibiotic-resistant strains.
Link to the study : https://www.biorxiv.org/content/10.1101/2024.05.28.596288v1.full.pdf