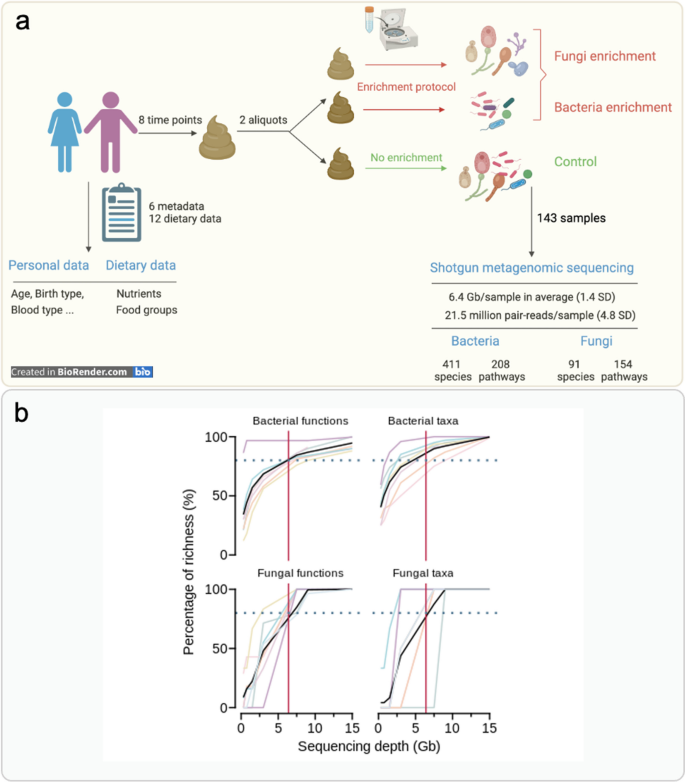
💡 This study presents a comprehensive analysis of the human gut microbiome, focusing on both bacterial and fungal communities, and explores their dynamic interactions influenced by dietary factors. Two sequencing approaches, internal-transcribed-spacer (ITS) and shotgun metagenomics, were compared to assess their accuracy in profiling the fungal community.
A novel fungal enrichment protocol was proposed to enhance the reliability of mycobiome analysis. The study also investigated the correlation between microbial communities and habitual diet composition.
📍 Methods:
📌 Sequencing Approaches Comparison: Utilized in silico simulations to assess the accuracy of ITS and shotgun metagenomics in mycobiome profiling. Implemented shotgun metagenomic sequencing with a fungal single-copy marker gene database for enhanced species-level resolution.
📌 Fungal Enrichment Protocol: Proposed a protocol to concentrate fungal cells in fecal samples. Employed centrifugation as a reliable method for fungal enrichment, comparing it with a membrane filter approach.
📌 Network Analysis: Conducted network analysis to identify keystone microbial species within the gut environment. Explored correlations between bacterial and fungal species, highlighting the potential role of certain species in maintaining gut microbiome balance.
📌 Dietary Correlation Analysis: Collected dietary data from study participants over a 2-month period. Investigated the relationship between microbial diversity, composition, and functions with habitual diet composition. Explored the impact of diet categories (sweets, protein, iron) on bacterial and fungal alpha diversity.
📍 Key Findings:
📌 Sequencing Approaches: Shotgun metagenomics demonstrated higher accuracy than ITS sequencing in mycobiome profiling at the species level. Advocated for the adoption of shotgun metagenomics for fungal microbiome studies due to its cost-effectiveness and ability to capture more comprehensive information.
📌 Fungal Enrichment Protocol: Proposed a fungal enrichment protocol using centrifugation, effectively increasing fungal counts and richness in fecal samples. Highlighted the limitations of membrane filter approaches in fungal enrichment.
📌 Network Analysis: Identified 𝘋𝘦𝘣𝘢𝘳𝘺𝘰𝘮𝘺𝘤𝘦𝘴 𝘩𝘢𝘯𝘴𝘦𝘯𝘪𝘪 as a keystone fungal species, potentially implicated in maintaining gut microbiome balance. Revealed inter-kingdom interactions between bacterial and fungal species, emphasizing the importance of studying both communities for a holistic understanding.
📌 Dietary Correlation Analysis: Discovered a dynamic association between microbial diversity, composition, and functions with habitual diet composition. Iron Impact: The study demonstrated, for the first time, a negative correlation between iron levels in the habitual diet and fungal functional alpha diversity, suggesting a potential competitive interaction between bacteria and fungi for iron-related resources.
📍 This study provides an efficient workflow for integrated bacterial and fungal gut microbiome profiling, shedding light on the dynamic interactions influenced by dietary factors. The findings emphasize the importance of studying both microbial kingdoms and their correlation with habitual diet for a comprehensive understanding of the human gut microbiome.
Read Further : https://tinyurl.com/msmktet5