💡 This study details the creation of #Twnbiome, a pioneering public gut microbiome database designed to facilitate microbiota research specifically for the Taiwanese population. The database offers a comprehensive overview of the gut microbiome composition in 119 healthy subjects from Taiwan.
Twnbiome not only provides a valuable baseline dataset for reference but also incorporates a user-friendly interface with an analysis pipeline, enabling non-bioinformatics users, such as clinicians and researchers, to conduct their own analyses effortlessly.
📍 Methods:
📌 Study Design and Participant Selection: The study included 119 healthy Taiwanese subjects recruited from the Health Management Center of National Taiwan University Hospital (NTUH). Informed consent was obtained, and participants underwent routine health checkups. Exclusion criteria were defined based on various factors, including disease history, lifestyle, and recent drug usage.
📌 Biosample Collections and 16S rRNA Sequencing: Fecal samples were collected, and DNA extraction was performed using the QIAamp DNA Stool Mini Kit. Sequencing of the V3 and V4 regions of the 16S rRNA gene was carried out using the Illumina HiSeq platform. Bioinformatic analysis involved quality control, sequence merging, filtering, operational taxonomic unit (OTU) identification, and taxonomic classification.
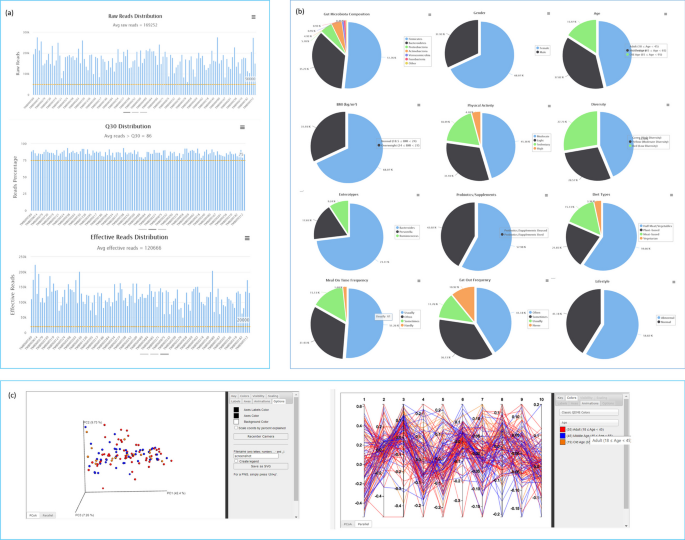
📌 Construction of Twnbiome Database: Twnbiome was developed using Django 2.2, Python 3.6.7, and MySQL 5.7.29. The database provides an overview of the gut microbiome composition, including demographic characteristics, quality control metrics, and beta diversity analysis.
📍 Key Findings:
📌 Demographic Characteristics: The study cohort comprised predominantly female participants (68.07%), with a mean age of 47.1 years. Subjects were categorized based on age, body mass index (BMI), physical activity, dietary habits, and lifestyle/sleep patterns.
📌 Data Analysis Tool: The Twnbiome database provides a data analysis tool where users can upload their own 16S rRNA sequence data. The analysis pipeline yields customizable plots and downloadable tables, allowing users to interpret their results effortlessly.
Twnbiome stands as a groundbreaking initiative in microbiome research, providing a comprehensive gut microbiome dataset for healthy Taiwanese individuals. The database not only serves as a reference control for case-control studies but also empowers non-bioinformatics users to conduct analyses through an intuitive interface. Twnbiome is poised to accelerate microbiota research in Taiwan, contributing valuable insights into the population-specific composition of the gut microbiome.
Link to the article : http://tinyurl.com/z5ywep7s