💡 Inflammatory bowel diseases (IBDs) are multifaceted disorders in which the immune system targets the gut microbiota in genetically predisposed individuals, triggered by largely unknown environmental factors.
📍 This study investigates metagenomic data from the gut microbiota of control subjects, Crohn’s disease (CD) patients, and ulcerative colitis (UC) patients. The data is analyzed by constructing correlation networks and co-expression networks, shedding light on the interconnectedness of the gut microbiota in these conditions.
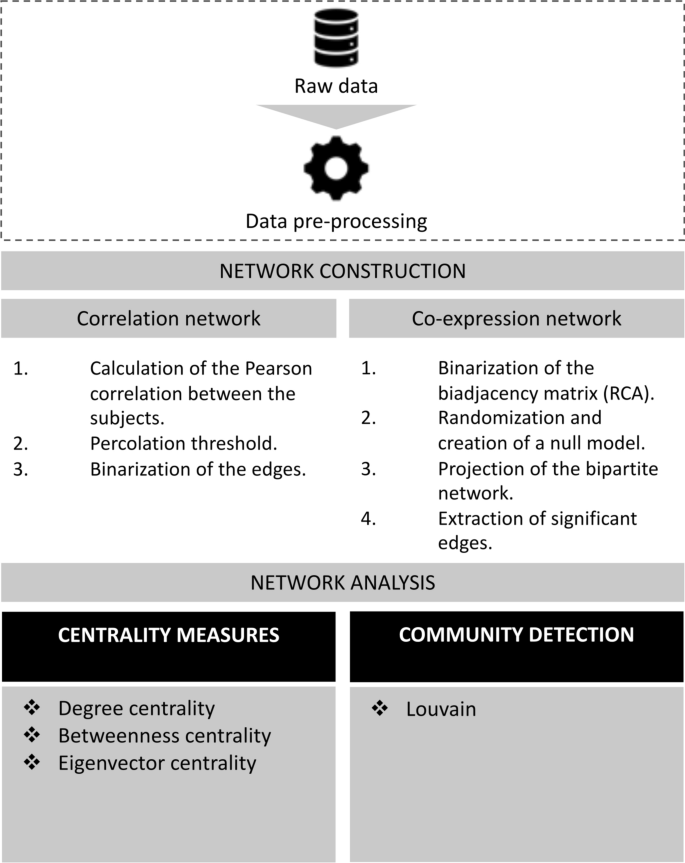
Methods:
📍 Correlation networks are created by calculating Pearson’s correlations between gene expression profiles across subjects. A percolation-based procedure is applied to threshold and binarize the adjacency matrices. Co-expression networks involve the construction of bipartite subjects-genes networks, followed by monopartite genes-genes projections after binarization of the biadjacency matrix. Centrality measures and community detection are employed to dissect the complexity of these networks and identify potential disease biomarkers.
Key Findings:
📌 Modules of Bacteroides, important gut bacteria, are interconnected in control subjects but isolated in IBD patients. This suggests that different Bacteroides species thrive independently in IBD, which aligns with previous findings associating lower Bacteroides levels with IBD.
📌 In IBD networks, 𝘍𝘢𝘦𝘤𝘢𝘭𝘪𝘣𝘢𝘤𝘵𝘦𝘳𝘪𝘶𝘮 𝘱𝘳𝘢𝘶𝘴𝘯𝘪𝘵𝘻𝘪𝘪, a critical bacterium, exhibits altered connectivity patterns, particularly in its co-enzyme A pathways. This deviation in connectivity hints at a change in the functionality of this bacterium rather than a decrease in quantity. Notably, coenzyme A is vital in fatty acid metabolism, previously linked to IBD.
📌 The 𝘌. 𝘤𝘰𝘭𝘪 module is observed in IBD, with varying interactions with other species in different diagnoses. The interaction of 𝘌. 𝘤𝘰𝘭𝘪 𝘸𝘪𝘵𝘩 𝘝. 𝘱𝘢𝘳𝘷𝘶𝘭𝘢 𝘢𝘯𝘥 𝘉. 𝘧𝘳𝘢𝘨𝘪𝘭𝘪𝘴 is influenced by pathways mediating these connections. Understanding the distinct wiring patterns may provide insights into IBD development.
📍 This network-based analysis uncovers complex alterations in gut microbiota interactions in IBD. Changes in connectivity patterns of key bacteria like 𝘍𝘢𝘦𝘤𝘢𝘭𝘪𝘣𝘢𝘤𝘵𝘦𝘳𝘪𝘶𝘮 𝘱𝘳𝘢𝘶𝘴𝘯𝘪𝘵𝘻𝘪𝘪 and the presence of distinct modules in different IBD types emphasize the intricacies of these diseases. Additionally, the roles of 𝘝. 𝘱𝘢𝘳𝘷𝘶𝘭𝘢 𝘢𝘯𝘥 𝘙. 𝘪𝘯𝘵𝘦𝘴𝘵𝘪𝘯𝘢𝘭𝘪𝘴 in the 𝘌. 𝘤𝘰𝘭𝘪 module provide potential avenues for further investigation into IBD mechanisms.
Link to the article : https://tinyurl.com/ycyauese