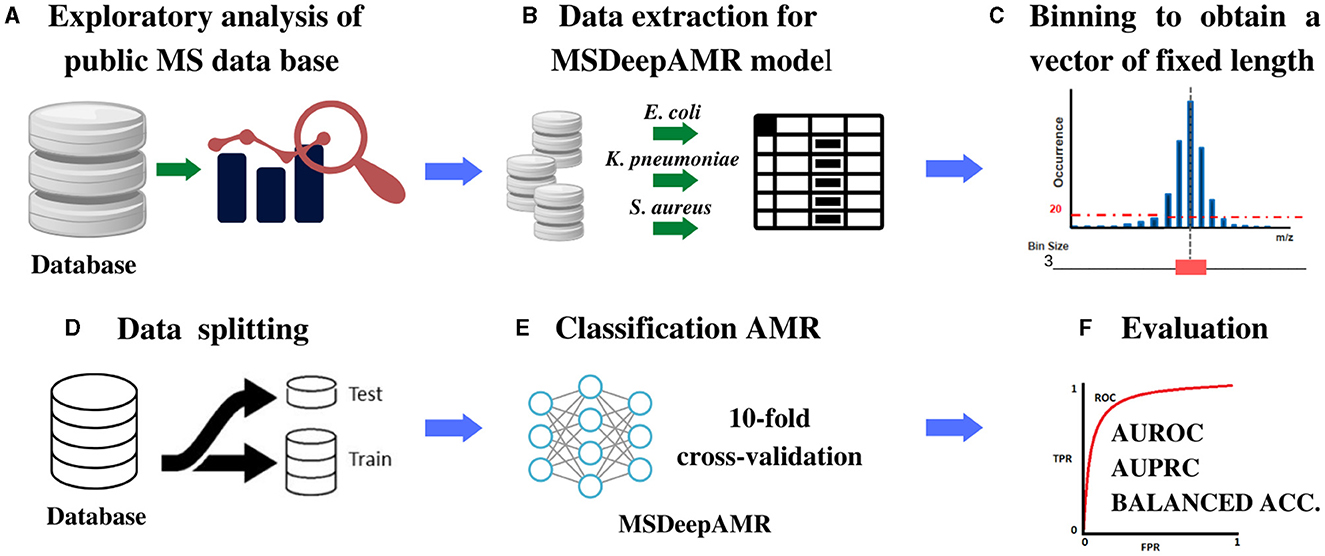
Antimicrobial resistance (AMR) poses a global health threat, necessitating early and accurate treatment strategies. Mass spectrometry (MS), particularly MALDI-TOF, is widely used in clinical microbiology to identify bacterial species and detect AMR. However, existing AMR prediction models often rely on manual preprocessing of spectra. This study introduces MSDeepAMR, a deep neural network trained on raw mass spectra to predict antibiotic resistance in 𝘌𝘴𝘤𝘩𝘦𝘳𝘪𝘤𝘩𝘪𝘢 𝘤𝘰𝘭𝘪, 𝘒𝘭𝘦𝘣𝘴𝘪𝘦𝘭𝘭𝘢 𝘱𝘯𝘦𝘶𝘮𝘰𝘯𝘪𝘢𝘦, 𝘢𝘯𝘥 𝘚𝘵𝘢𝘱𝘩𝘺𝘭𝘰𝘤𝘰𝘤𝘤𝘶𝘴 𝘢𝘶𝘳𝘦𝘶𝘴.
Key Scientific Findings
- MSDeepAMR Model Performance:
- MSDeepAMR demonstrated robust classification performance for antibiotic resistance, with AUROC values consistently above 0.83 across various bacterial species and antibiotics.
- Compared to previous investigations, MSDeepAMR improved AUROC results by over 10%, showcasing its superior performance in AMR prediction.
- Transfer Learning Benefits:
- Transfer learning experiments indicated the adaptability of pre-trained MSDeepAMR models to external data, resulting in AUROC improvements of up to 20% compared to models trained solely on external data.
- Despite challenges in adapting models to new laboratory data, retraining all layers of a pre-trained model proved more effective than training from scratch.
- Comparison with State-of-the-Art:
- MSDeepAMR outperformed traditional machine learning algorithms by an average of 13% in AUROC values and showed considerable improvement in AUPRC, doubling or even tripling previous results in some cases.
- Normalization and regularization layers were found to enhance model performance, particularly in addressing class imbalance issues in datasets.
- Implications for Clinical Practice:
- MSDeepAMR offers a complete methodology for AMR prediction from raw mass spectrometry data, leveraging deep learning to identify complex patterns.
- The model’s high performance and adaptability make it a promising tool for clinical laboratories, enabling accurate and efficient AMR detection even with limited sample collections.
- Transfer learning facilitates the reproducibility of MSDeepAMR models across different laboratories, ensuring their utility in diverse clinical settings.
MSDeepAMR represents a significant advancement in AMR prediction, leveraging deep learning to analyze raw mass spectra for accurate classification of antibiotic resistance. Its superior performance, demonstrated adaptability through transfer learning, and potential for on-the-fly detection make it a promising tool for clinical microbiology. With ongoing optimization and collaboration to build extensive databases, MSDeepAMR holds great potential in combatting the growing threat of antimicrobial resistance worldwide.
Link to the article : https://tinyurl.com/sbdt6b66