💡 The association between gut microbiome and schizophrenia spectrum disorders (SSD) remains underexplored. In this study, fecal shotgun metagenomic sequencing was utilized to analyze bacteria and bacterial genes in 52 young adult SSD patients and 52 controls. Compared to controls, young SSD patients exhibited significantly lower α-diversity and distinct β-diversity in both bacterial species and functional genes. Notably, machine learning classifiers achieved an accuracy of 70% in classifying SSD individuals based on their gut microbiome profiles. Differential abundance analysis revealed higher abundance of oral cavity-associated species and amino acid biosynthesis modules in SSD patients, while butyrate synthesis and acetate degradation modules were less abundant. The findings underscore the presence of gut microbiome alterations in SSD and highlight the potential of machine learning algorithms for patient classification.
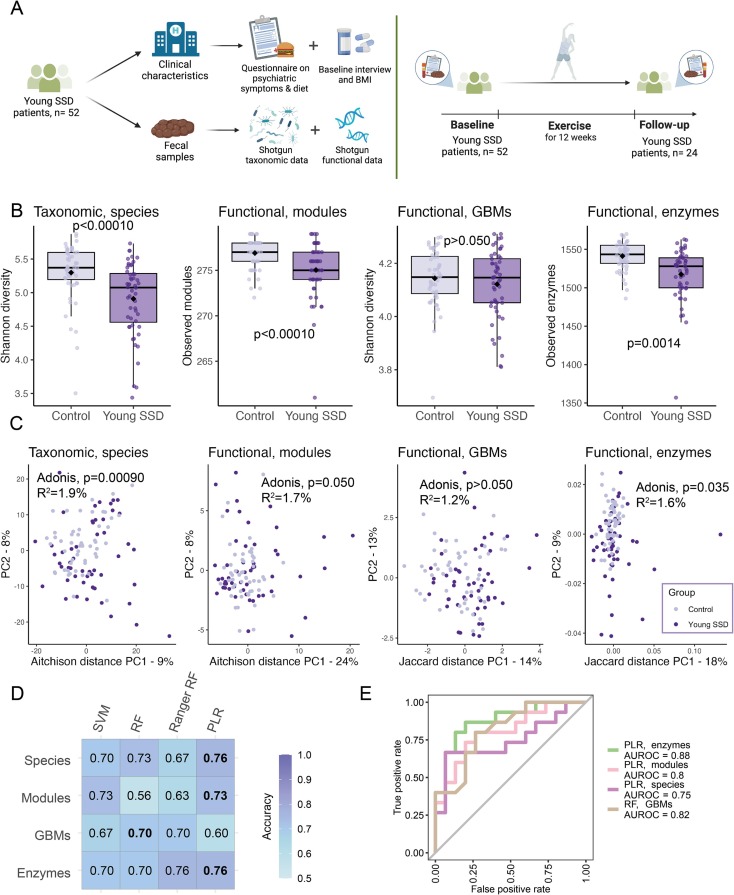
📍 Case-Control Microbiome Diversity: Young SSD patients demonstrated lower α-diversity in bacterial species and functional genes compared to controls, independent of age, sex, BMI, and diet intake.
β-diversity analysis revealed significant differences in bacterial and functional diversity metrics between SSD patients and controls, suggesting distinct gut microbiome profiles.
📍 Case-Control Classification and Differentially Abundant Microbiome Features: Machine learning classifiers achieved 76% accuracy in distinguishing SSD patients from controls based on taxonomic and functional data.
Differential abundance analysis identified oral cavity-associated species and amino acid biosynthesis modules as enriched in SSD patients, while butyrate synthesis and acetate degradation modules were depleted.
📍 Antipsychotic Use and Microbiome Diversity: Nine SSD patients not taking antipsychotics exhibited lower evenness compared to controls, suggesting an association between antipsychotic medication and microbiome diversity.
This study reveals significant alterations in the gut microbiome of young adults with SSD, characterized by reduced α-diversity, distinct β-diversity, and differential abundance of oral cavity-associated species and amino acid biosynthesis modules. Machine learning classifiers demonstrate potential for patient classification based on microbiome profiles. Understanding these microbiome changes may offer insights into SSD pathophysiology and facilitate the development of targeted interventions. Further research is warranted to validate these findings and elucidate underlying mechanisms.
Link to the article : http://tinyurl.com/377fda64